By accessing this website we assume you accept these terms and conditions. Do not continue to use Int&in if you do not agree to take all of the terms and conditions stated on this page. The following terminology applies to these Terms and Conditions, Privacy Statement and Disclaimer Notice and all Agreements: "Client", "User", "You" and "Your" refers to you, the person log on this website and compliant to the Company's terms and conditions. "The Company", "Ourselves", "We", "Our" and "Us", refers to our Company. "Party", "Parties", or "Us", refers to both the Client and ourselves. All terms refer to the offer, acceptance and consideration of payment necessary to undertake the process of our assistance to the Client in the most appropriate manner for the express purpose of meeting the Client's needs in respect of provision of the Company's stated services, in accordance with and subject to, prevailing law of Netherlands. Any use of the above terminology or other words in the singular, plural, capitalization and/or he/she or they, are taken as interchangeable and therefore as referring to same.
Cookies
We employ the use of cookies. By accessing Int&in, you agreed to use cookies in agreement with the Int&in's Privacy Policy. Most interactive websites use cookies to let us retrieve the user's details for each visit. Cookies are used by our website to enable the functionality of certain areas to make it easier for people visiting our website.
License
Parts of this website offer an opportunity for users to upload documents. Int&in does not filter, edit, publish or review these documents prior to their presence on the website. To the extent permitted by applicable laws, Int&in shall not be liable for the documents or for any liability, damages or expenses caused and/or suffered as a result of any use of and/or uploading of and/or appearance of the documents on this website.
Int&in reserves the right to monitor all documents and to remove any documents should we deem it necessary.
You warrant and represent that:
1) You are entitled to upload the documents on our website and have all necessary licenses and consents to do so;
2) The documents do not invade any intellectual property right, including without limitation copyright, patent or trademark of any third party;
3) The documents do not contain any defamatory, libelous, offensive, indecent or otherwise unlawful material which is an invasion of privacy
4) The documents will not be used to solicit or promote business or custom or present commercial activities or unlawful activity.
You hereby grant Int&in a non-exclusive license to host and monitor any documents you upload on this site.
Further you agree that any uploaded documents are accessible through a randomly generated unique key for each individually uploaded document.
Commercial use
Commercial use is not permitted. The web server is free to use for academic purposes.
Hyperlinking to our Content
You may link to our web site so long as the link:
(a) is not in any way deceptive;
(b) does not falsely imply sponsorship, endorsement or approval of the linking party and its products and/or services; and
(c) fits within the context of the linking party's site.
Reservation of Rights
We reserve the right to request that you remove all links or any particular link to our Website. You approve to immediately remove all links to our Website upon request. We also reserve the right to amen these terms and conditions and it's linking policy at any time. By continuously linking to our Website, you agree to be bound to and follow these linking terms and conditions.
Removal of links from our website If you find any link on our Website that is offensive for any reason, you are free to contact and inform us any moment. We will consider requests to remove links but we are not obligated to or so or to respond to you directly. We do not ensure that the information on this website is correct, we do not warrant its completeness or accuracy; nor do we promise to ensure that the website remains available or that the material on the website is kept up to date.
Disclaimer
To the maximum extent permitted by applicable law, we exclude all representations, warranties and conditions relating to our website and the use of this website. Nothing in this disclaimer will:
1) limit or exclude our or your liability for death or personal injury;
2) limit or exclude our or your liability for fraud or fraudulent misrepresentation;
3) limit any of our or your liabilities in any way that is not permitted under applicable law; or
4) exclude any of our or your liabilities that may not be excluded under applicable law.
The limitations and prohibitions of liability set in this Section and elsewhere in this disclaimer: (a) are subject to the preceding paragraph; and (b) govern all liabilities arising under the disclaimer, including liabilities arising in contract, in tort and for breach of statutory duty.
As long as the website and the information and services on the website are provided free of charge, we will not be liable for any loss or damage of any nature.
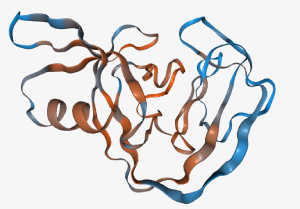 The structure is colored according to the probability that the split sites are active (blue) and inactive (red).
The structure is colored according to the probability that the split sites are active (blue) and inactive (red).
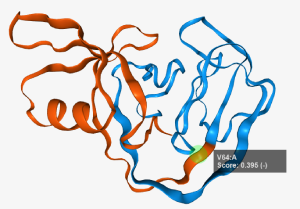 Residues can be selected via Ctrl+Leftclick (a green sphere will show the selection) which treat the selected residue as the site where the protein is split. The split occurs after the selected residue.
Additionally the two split fragments are shown with two colors (red and blue).
Selected residues can be deselected via Ctrl-Leftclicking anywhere but on a residue. A tooltip will appear when
hovering the mouse over the residue. The residues' chain information is displayed after a colon.
Residues can be selected via Ctrl+Leftclick (a green sphere will show the selection) which treat the selected residue as the site where the protein is split. The split occurs after the selected residue.
Additionally the two split fragments are shown with two colors (red and blue).
Selected residues can be deselected via Ctrl-Leftclicking anywhere but on a residue. A tooltip will appear when
hovering the mouse over the residue. The residues' chain information is displayed after a colon.
 The current camera view can be saved via this button.
The current camera view can be saved via this button. The size of the structure window can be resized via this button.
The size of the structure window can be resized via this button.
 Different groups of split sites can be selected outputting a list of split sites ordered from the highest activity probability to lowest.
The high active probability group encompasses sites with a activity probability of at least 0.75. The high inactive probability encompasses sites with a activity probability of less than 0.25.
The active and inactive groups have all sites that the model would predict to be active (at least 0.5) and inactive (less than 0.5) respectively.
Different groups of split sites can be selected outputting a list of split sites ordered from the highest activity probability to lowest.
The high active probability group encompasses sites with a activity probability of at least 0.75. The high inactive probability encompasses sites with a activity probability of less than 0.25.
The active and inactive groups have all sites that the model would predict to be active (at least 0.5) and inactive (less than 0.5) respectively.
 The list can be copied to the clipboard with this button.
The list can be copied to the clipboard with this button.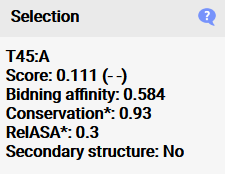 When a residue is selected, additional information concerning the conservation,
binding affinity, surface accessibility and secondary structure properties are shown here which are used to calculate the activity probability in the model. It is important to note that
conservation and surface accessibility are given as means of the surrounding residues of the split site, as this was shown to work best for predicting the activity,
so the values do not show single residue values (which can be accessed in the raw files) (*). The split site is considered to be in a secondary structure if both flanking residues of the split site are in
a secondary structure region.
When a residue is selected, additional information concerning the conservation,
binding affinity, surface accessibility and secondary structure properties are shown here which are used to calculate the activity probability in the model. It is important to note that
conservation and surface accessibility are given as means of the surrounding residues of the split site, as this was shown to work best for predicting the activity,
so the values do not show single residue values (which can be accessed in the raw files) (*). The split site is considered to be in a secondary structure if both flanking residues of the split site are in
a secondary structure region.
 Deselecting residues can be done in the structure view or with this button located at the top of the Data tab.
Deselecting residues can be done in the structure view or with this button located at the top of the Data tab.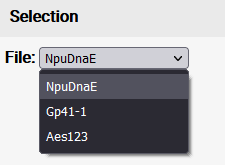 If multiple files were uploaded via batch upload, the single files can be selected via a dropdown menu.
If multiple files were uploaded via batch upload, the single files can be selected via a dropdown menu. Zooming out can be done via a double-click on the graph or via this button that appears at the top-left of the graph (if it is zoomed in).
Zooming out can be done via a double-click on the graph or via this button that appears at the top-left of the graph (if it is zoomed in).
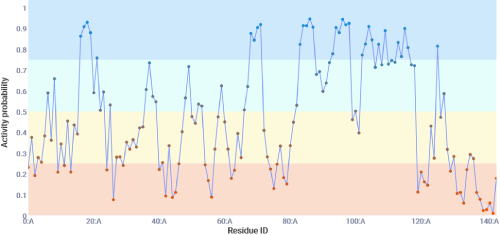 This tab shows the models prediction for each possible split site. There are 4 categories given with 4 colors.
Generally speaking the model predicts split sites with a probability of at least 0.5 to be active and below 0.5 to be inactive. Sites with a probability of at least 0.6 have been shown to
contain the highest ratio of true positives to false positives at the dispense of loosing some true positive split sites. Sites with a probability of less than 0.4 have been shown to
contain the highest ratio of true negatives to false negatives.
This tab shows the models prediction for each possible split site. There are 4 categories given with 4 colors.
Generally speaking the model predicts split sites with a probability of at least 0.5 to be active and below 0.5 to be inactive. Sites with a probability of at least 0.6 have been shown to
contain the highest ratio of true positives to false positives at the dispense of loosing some true positive split sites. Sites with a probability of less than 0.4 have been shown to
contain the highest ratio of true negatives to false negatives.
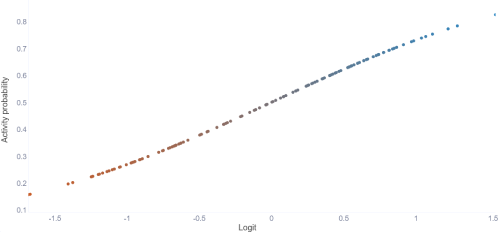 This tab shows the logit function, which what the model's prediction is based on. The points are sorted by the lowest prediction to the highest (Meaning the splits are not in order like with the other tabs).
This tab shows the logit function, which what the model's prediction is based on. The points are sorted by the lowest prediction to the highest (Meaning the splits are not in order like with the other tabs).
 The sequence tab shows the sequence with the activity probability color codes,
residue id's and whether the residue belongs to a secondary structure region (marked with Sec).
The sequence tab shows the sequence with the activity probability color codes,
residue id's and whether the residue belongs to a secondary structure region (marked with Sec).
 The raw data can be downloaded in this tab as a zip file.
The zip contains the output from DSSP, hmmer, muscle and rate4site as well as the pdb file that was generated through pdb2pqr at a pH of 7.
The raw data can be downloaded in this tab as a zip file.
The zip contains the output from DSSP, hmmer, muscle and rate4site as well as the pdb file that was generated through pdb2pqr at a pH of 7.